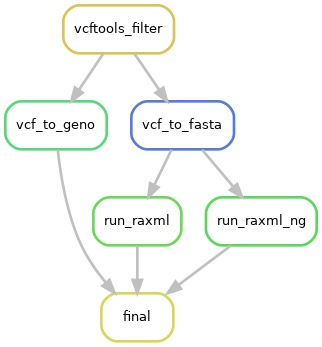
How used the best SNP calling workflow for GBS or full genome sequencing !!!!
From fastq to bam
RattleSNP is a really flexible tool mapped paired data. You can give parameters on the config.yaml file to RattleSNP to generate a modular, dedicated pipeline for your own data.
Mapping
RattleSNP includes (at the moment) only BWA tools.
Included tools :
Optional statistics
Using RattleSNP you can activate or deactivate statistics on mapping steps.
Directed acyclic graphs (DAGs) show the differences between deactivated (BUILD_STATS=False):
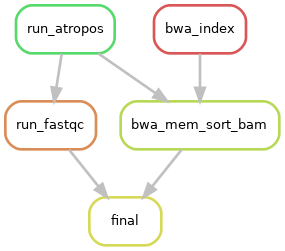
and activated BUILD_STATS step on configuration file (BUILD_STATS=True):
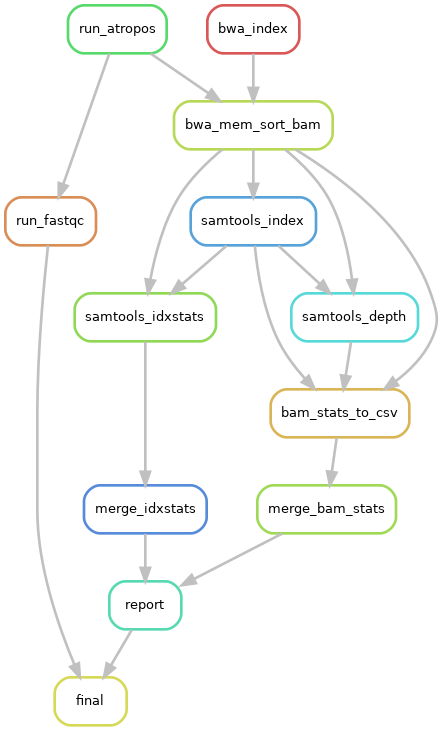
Warning
In any case, MAPPING will be activated if BUILD_STATS is True even if MAPPING-ACTIVATE is False
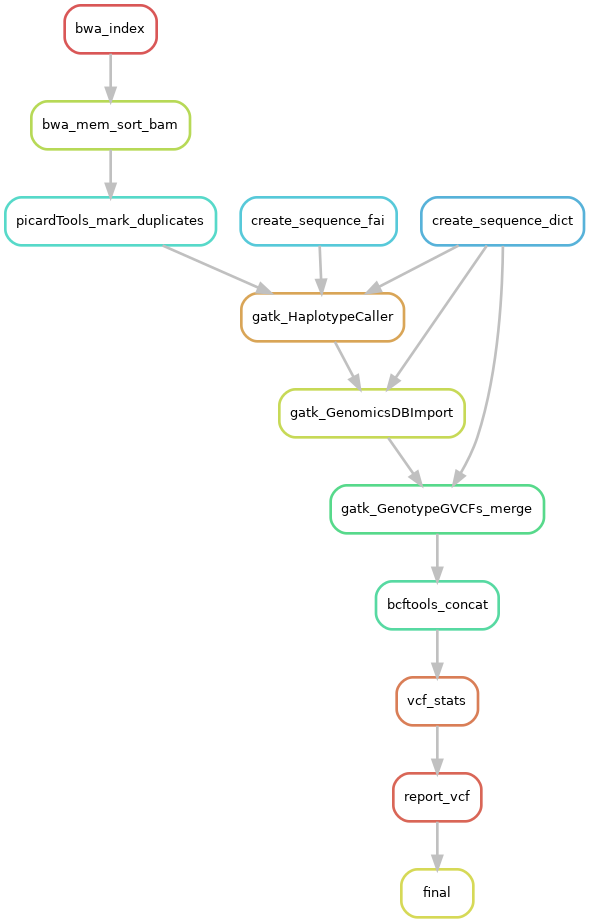
SNP calling
The SNP calling is perform by GATK tools.
Included tools :
GATK version >= 4.0.0 https://gatk.broadinstitute.org/hc/en-us/articles/360036194592-Getting-started-with-GATK4
Filter VCF and genetics tools